From R to Python with minimal baggage
Getting the best of both worlds.
Navigating between R and Python can appear intimidating. Yet, once the right strategy is found, interoperability becomes a stroll through the sunshine.
For this blog post, I spoke with Luke Zappia, Philipp Angerer, and Tomasz Kalinowski, who have experienced R and Python interoperability from both user and developer perspectives. Here, I share the aspects they identified as indispensable for a smooth interoperability journey. The two red threads of our conversations were making the interoperability user-friendly and efficient.
Luke is a bioinformatics scientist who specializes in benchmarking, reproducible workflows, and interoperability. He is also the author of the zellkonverter R package, which converts between R and Python objects for storing single-cell measurement data.
Philipp is a software developer who has contributed to various open-source packages across multiple programming languages. He is also the author of the anndata2ri package, which serves a similar function to zellkonverter but is designed for Python.
Tomasz is a core developer of the reticulate package, which facilitates Python interoperability from within R. Besides, he enables R users easy access to deep learning tools, including TensorFlow and Keras.
Although many examples in this blog post focus on the R-to-Python direction, the discussed principles often translate to the reverse direction.
While this blog post is based on discussions with Luke, Philipp, and Tomasz, I often omit their names for the sake of readability.
Sections list
- The flavors of interoperability
- Reasons to try out interoperability
- Learning from other languages
- Breaking down perceptual walls
- Starting in the right environment
- Transferring information between R and Python
- The curse of custom data formats
- Limits of converting objects
- Efficient conversion is no conversion
- Details on sharing memory between R and Python
- From propagating errors to propagating error messages
- Managing processes
The flavors of interoperability
In the past, switching between R and Python relied on saving data in one language and reading it into the other (Figure 1). This method works reasonably well for automated pipelines that run a series of scripts on a server, like Snakemake. However, it is inconvenient for interactive workflows where one wishes to run the entire analysis within a single notebook.
Since then, packages that allow direct access to the functionality of one language while working in the other language have been established (Figure 1). These packages enable in-memory transfer of objects between languages and provide an interface to functions from the other language. The most notable packages are reticulate, for calling Python code from R, and rpy2 for calling R code from Python.
Lately, the growing dataset sizes have induced another workflow shift. It is becoming common to keep the data on disk and load into memory only parts required for computation, known as out-of-core computation1. Philipp pointed out that this trend might influence how interoperability packages develop in the future (Figure 1).
Moving from in-memory to out-of-core interoperability
As datasets grow, relying on traditional in-memory operations for the entire dataset is no longer possible. Instead, a more efficient approach involves loading datasets in chunks and performing operations on each chunk separately before combining the results. This decreases memory requirements and enables parallelization for faster computation.
With data stored on disk instead of in memory and loaded on demand, the need for transferring in-memory objects between languages is reduced. However, to facilitate out-of-core interoperability, frameworks that can efficiently handle these data formats need to be more widely adopted in both languages.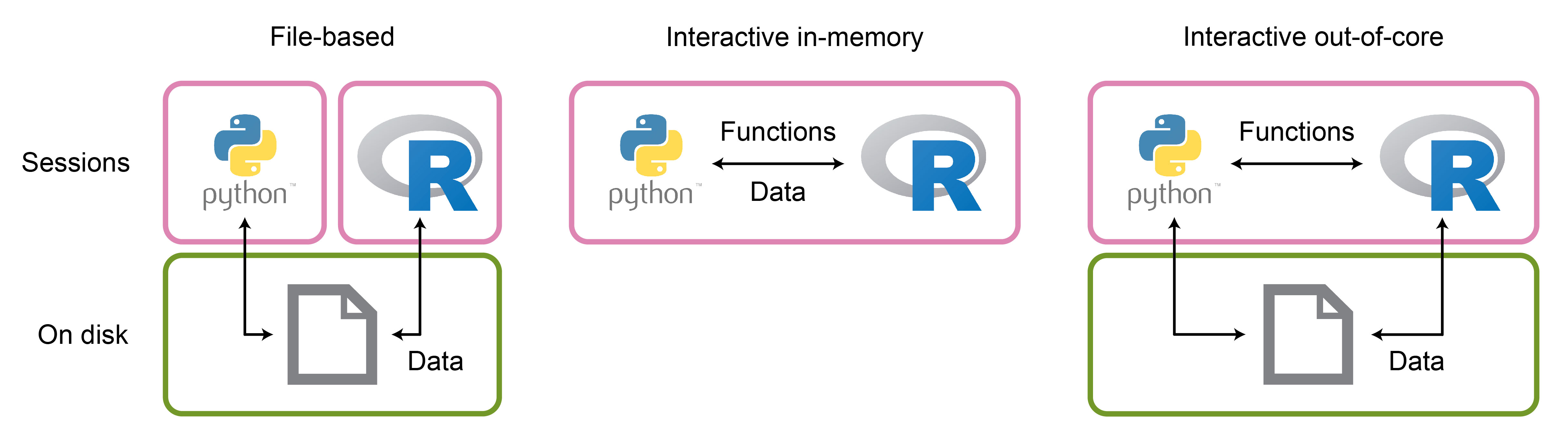 Figure 1: Different interoperability approaches. The term “interoperability” in the context of R and Python usually refers to interactive in-memory interoperability via reticulate or rpy2.
Figure 1: Different interoperability approaches. The term “interoperability” in the context of R and Python usually refers to interactive in-memory interoperability via reticulate or rpy2.
Backend interoperability
Apart from user-facing interoperability, interoperability is also widely employed in package development. For example, the backends of R and Python packages are often implemented in high-performance languages, such as C. Besides enhancing performance, this eliminates redundant work. Namely, to provide equivalent functionality in another language, only the binders that provide user access to the backend must be developed, as seen in igraph.
Recently, Rust has gained traction as a backend language thanks to its high-performance nature combined with integrated interoperability functions for several languages. Accordingly, Philipp anticipates that the popularity of Rust backends will continue to grow.Reasons to try out interoperability
If a solution already exists in one language, there may be little reason to reinvent the wheel in another language. Often, interoperability can enable new functionalities at a lower cost.
For instance, R offers many packages tailored to specific statistical or data science tasks. Although these problems can be solved in Python with general statistical frameworks like statsmodels, the code may be significantly more complex. Moreover, since these specialized packages often have only small communities, recreating them in another language might not be the best investment of developer time.
Another reason for tapping into a different language are natively provided complementary functionalities. For example, to efficiently map elements to keys for future retrieval (i.e., hashmaps), R requires specialized libraries, such as fastmap or collections. In contrast, Python provides this functionality natively via dictionaries. Similarly, while R vectors are copied when adding elements, some Python data structures allow adding elements without copying. Thus, Tomasz pointed out that in some cases2 Python, especially its standard library, can replace functionalities missing in R. On the other hand, R’s pipe functionality can be used to chain Python data operations, making the code more concise and readable2.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
## Using Python dictionary in R as an efficient hashmap
> library(reticulate)
> d=dict()
> d['a']=1
> d[0]=2
> d
{'a': 1.0, 0.0: 2.0}
> class(d)
[1] "python.builtin.dict" "python.builtin.object"
## Using Python deque in R as a memory-efficient vector alternative
# deque is a list-like object with fast appending and popping on both ends
# Create a deque and inspect its memory location
> deque <- reticulate::import("collections")$deque(c(1,2))
> deque
deque([1.0, 2.0])
> .Internal(inspect(deque))
@11fd633f8 04 ENVSXP g0c0 [OBJ,REF(1),ATT] <0x11fd633f8>
# After removing an object from deque, its memory location remains the same
> deque$pop()
[1] 2
> deque
deque([1.0])
> .Internal(inspect(deque))
@11fd633f8 04 ENVSXP g0c0 [OBJ,REF(1),ATT] <0x11fd633f8>
# Create an R vector and inspect its memory location
> v<-c(1,2)
> v
[1] 1 2
> .Internal(inspect(v))
@10d861a48 14 REALSXP g0c2 [REF(1)] (len=2, tl=0) 1,2
# When removing an object from a vector, a new vector is created
> v[-length(v)]
[1] 1
> .Internal(inspect(v[-length(v)]))
@10db46028 14 REALSXP g0c1 [] (len=1, tl=0) 1
Limitations of R’s inbuilt maps
Although R provides native hashmaps, known as environments, they come with several drawbacks. Environments can lead to memory leaks since their keys are R symbols, which are not subject to garbage collection. This undermines the primary purpose of hashmaps, which is to provide efficient access to many elements via unique keys. Moreover, keys must be strings. This limits their use when keys are dynamically generated and cannot be guaranteed to be character-based, requiring conversion. Such conversion can lead to clashes, e.g., the string ”FALSE” and the boolean FALSE would be converted to the same character representation.
Learning from other languages
Interaction with multiple programming languages naturally broadens one’s coding repertoire. This can be well illustrated by the language Haskell. While it is not commonly used in practice, many people have sharpened their recursion skills through it. In Haskell, recursion is the only way to perform iterative operations that rely on for and while loops in other languages. Once learned, recursion can also be useful for specific problems in other languages, such as tree parsing.
One reason behind the popularity of R in the applied data science community is its user-friendliness. For example, documentation is not only straightforward to create in R but is also “culturally” expected. Motivated by this, Philipp strives to replicate R’s approachable user experience in his software packages. He highly values meaningful error messages that help users resolve problems according to the “fail early, fail loudly” philosophy. Additionally, he advocates for easily readable printed-out messages, for which he recommended the terminal output formatting package rich.
Why should functions fail early and loudly?
NaN or infinite outputs. Even if this does not immediately trigger an error, such values can cause problems later in the chain of operations. As a result, diagnosing the source of error becomes difficult, since the stack trace may not be directly linked to the operation that initially created the NaN or infinite values. To prevent this, developers should implement safeguards against passing invalid input parameters.On the other hand, Philipp mentioned that well-written Python packages can serve as valuable references for structuring complex code bases. In Python, packages often consist of hierarchically organized namespaces (i.e., modules), which group objects based on their function. In contrast, R by default only supports flat namespaces (Figure 2). Good namespace organization benefits users and developers alike by providing a high-level functionality overview and as a safeguard against naming collisions, especially in large code bases.
Benefits of well-organized namespaces
Hierarchical namespaces provide a mental framework for understanding the functionality offered by the package for both developers and users. For example, R API documentation is typically arranged alphabetically because of its flat internal structure. In contrast, Python API documentation is usually organized based on functional modules. This allows users to gain an overview of the package's high-level capabilities before exploring individual objects within a specific module.
Moreover, namespace organization helps prevent name clashes. A common problem is the occurrence of namespace clashes when importing packages in R. Specifically, in R, it is common to import all object names from a package directly into the working environment. This can create clashes when multiple packages share the same names, and sometimes these conflicts may remain undetected, causing names to be unexpectedly overwritten. In contrast, in Python, it is common to preserve package and module information during imports, effectively preventing name clashes.
To alleviate these issues in R, different strategies for detecting and resolving clashes were introduced. For example, the package conflicted was created to assist in detecting and resolving conflicts. Moreover, to prevent conflicts from arising, the package::function notation allows users to specify the exact package from which to call a function. Import management was further expanded by the import and box packages. Like Python, these packages allow users to import specific names, rename them, and even maintain the source of the import (e.g., package) or group functions into distinct naming environments, which can be accessed as package$function or environment$function.
Furthermore, hierarchically organizing namespaces can help manage clashes during package development. For example, when creating a package for numerical operations, we might define two distinct sets of functions: one for scalars and another for matrices. In Python, this could be accomplished by defining two modules called scalar and matrix, each containing a dedicated multiply operation. The two would then be accessed as package.scalar.multiply and package.matrix.multiply. Since R does not support modules, we would need to define two functions with distinct names, such as multiply_scalar and multiply_matrix. Another alternative is to use S3 generics, which enable the overloading of generic functions (e.g., multiply). In this case, the types of the input parameters dictate which version of the overloaded function will be executed (e.g., multiply developed for scalars or matrices). However, this strategy is applicable only if the input types are distinct.
scalar or matrix) and stay bound to its scope without interference from other parts of the package. Moreover, we can define private module functions, which will not be used outside the specific module. In contrast, in R, we must always keep track of all names within a package, which becomes tedious to mentally track as the package expands. Consequently, there have been attempts to modularize R code (e.g., via box and modules packages); however, they are not as widely adopted and mature as in Python.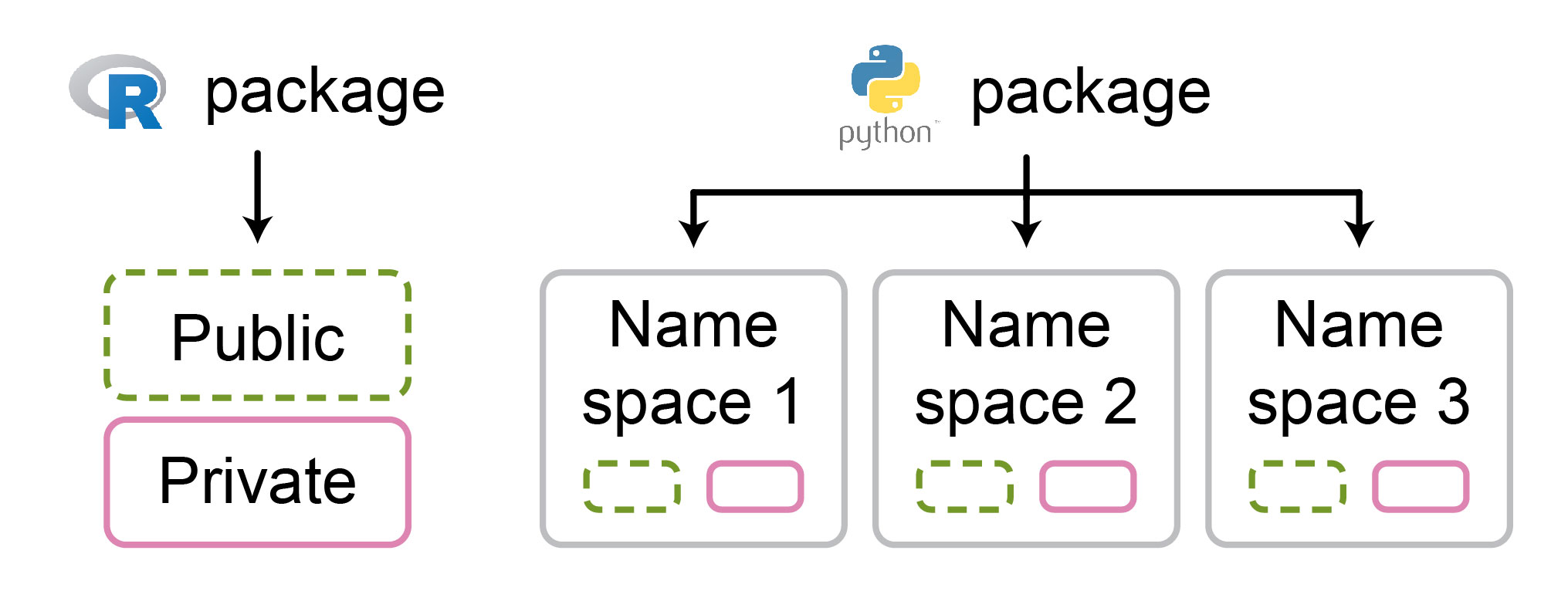 Figure 2: R packages have flat namespaces, while in Python packages, the namespaces can be hierarchically organized.
Figure 2: R packages have flat namespaces, while in Python packages, the namespaces can be hierarchically organized.
Accessibility of packaging to newcomers
Breaking down perceptual walls
Clearly, using R and Python together has many practical benefits. So why don’t we see it more frequently in practice? Luke pointed out that the main obstacle is usually not technical. From his experience, it is harder to persuade users that tapping into another language is worth the effort.
A common concern among native R users is that Python feels inaccessible. While the R community is known for warmly welcoming newcomers without a computer science background, the Python community can sometimes seem more intimidating. However, Tomasz explained that this perception is oversimplified. - The Python community is very diverse, and within it, many sub-communities foster the same atmosphere as in R.
For example, R is renowned for its vignettes, but these days, many Python packages also offer tutorials that guide users on integrating the API into a complete analysis pipeline. In many cases, functionality from another language can be used simply by following a tutorial, without first diving into its syntax. This is what makes interoperability tools so valuable; we can handle data preparation in a language we are comfortable with and then step into another language just to call a specific function.
On the other hand, Luke raised the issue of Python users often being dismissive towards the R language due to its perceived inefficiency and peculiar syntax. Here, I must admit that I am also one of those Python users who get shivers running down their spine when seeing R code. However, over the years of doing data science in Python while staying connected with the R community, I have come to realize that some tasks are just better to be done in R. For example, I often use heatmaps from the Python package seaborn3 for my exploratory data analysis, and they “do their job”. Nevertheless, I could not write a single paper without relying on the R package ComplexHeatmap (Figure 3) in combination with rpy2 to make the heatmaps look presentable.
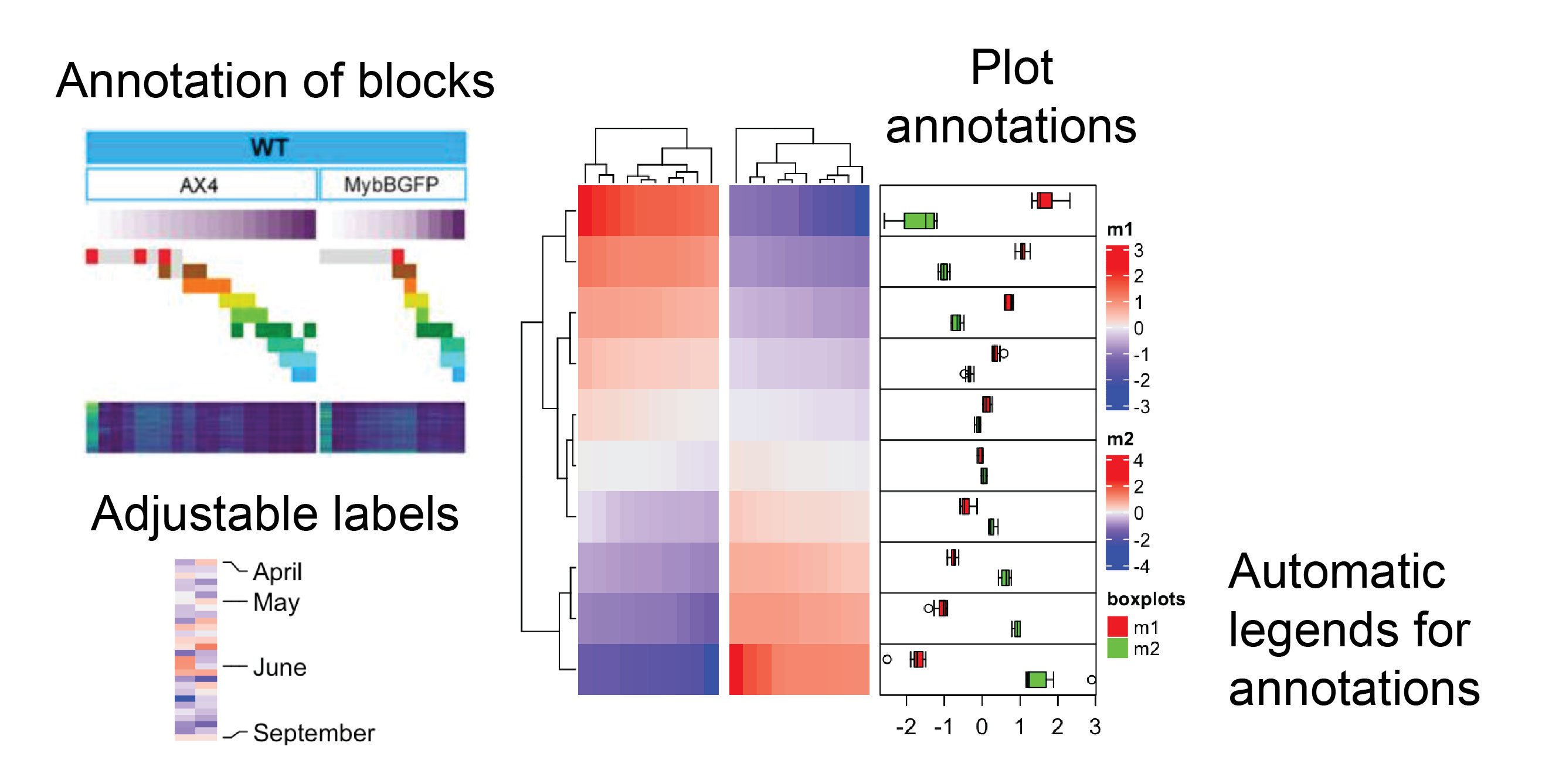 Figure 3: Examples of heatmap plotting capabilities from the ComplexHeatmap R package that are missing from the seaborn Python package. The plots were adapted from ComplexHeatmap documentation, which is under CC0-1.0 license, and one of my papers, which is under CC BY-NC 4.0 license.
Figure 3: Examples of heatmap plotting capabilities from the ComplexHeatmap R package that are missing from the seaborn Python package. The plots were adapted from ComplexHeatmap documentation, which is under CC0-1.0 license, and one of my papers, which is under CC BY-NC 4.0 license.
Starting in the right environment
While reticulate and rpy2 are not hard to use, they can be challenging to set up. Users must manage environments for both R and Python, and often these are not properly discovered by the other language. In response to this issue, the developers of reticulate undertook the challenge of developing a solution that smooths the setup process.
In the past, reticulate provided multiple options for configuring Python environments through conda and venv. However, Python is not unfairly notorious for its dependency conflicts, presenting a daunting hurdle, especially for new users. In comparison, the deployment of R packages is often more strictly regulated via initiatives like Bioconductor4. Thus, developers have higher awareness of conflicts related to their packages; hence, the burden is shifted away from the users.
Recently, uv transformed the installation of Python packages with its speed, allowing for on-the-fly installation (Figure 4). This is a significant shift from the traditional method of maintaining more widely-purposed environments, which tend to accumulate many packages with pinned dependency versions over time, often leading to conflicts. In contrast, with uv, it is possible to quickly create a fresh environment for every coding session5, reducing the chance of conflicts. Furthermore, uv outputs user-friendly messages upon dependency conflicts, simplifying manual resolution.
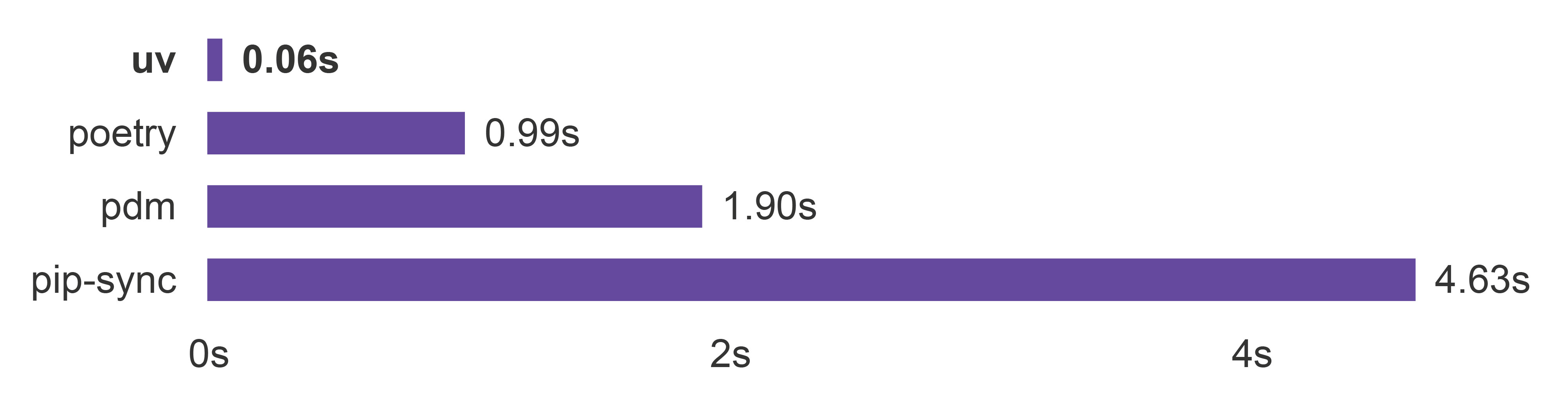 Figure 4: Time required to install dependencies of an example Python package (Trio) with a warm cache. A warm cache means that package data has been previously downloaded. The figure was adapted from uv’s GitHub page, which is under MIT or Apache-2.0 license.
Figure 4: Time required to install dependencies of an example Python package (Trio) with a warm cache. A warm cache means that package data has been previously downloaded. The figure was adapted from uv’s GitHub page, which is under MIT or Apache-2.0 license.
Transferring information between R and Python
After setting up the interoperability environments, the next obstacle the users hit is data transfer between the two languages. While a bit of practice can bring one a long way, additional considerations are required to make the process truly efficient.
The curse of custom data formats
The ease of interoperability largely hinges on how straightforward it is to convert data between languages. To help with this, both reticulate and rpy2 provide converters for built-in data structures, such as lists, and object classes from popular packages, such as NumPy. However, users and developers must define conversion rules for other objects.
Unfortunately, applied data science communities often rely on specialized data formats, complicating conversion. To make matters worse, the formats for individual data types (e.g., gene expression measurements) also differ within individual languages.
This disparity is driven by multiple factors. Often, data is stored in custom formats to simplify the specialized analyses provided by individual packages. Moreover, before a standard format for a specific data type had been established, multiple independent formats may have emerged from concurrently developed packages. However, it is also often the case that developers decide not to use the existing infrastructure. For example, since academic work is usually evaluated based on citation counts, it may be less rewarding to employ a package managed by another group than to promote one’s own repositories. Furthermore, it is often easier to publish a paper with a new package rather than by maintaining an old one, leading to many stale packages. However, to make a package user-friendly, it is important to consider how it integrates into the broader ecosystem.
Limits of converting objects
To convert objects between R and Python, it is important to not only understand how those objects are defined in the source language, but also how the two languages differ in representing them6.
Philipp pointed out that some data types are unique to individual languages. For instance, in Python, integers can grow without limitations, whereas in R, the native integer type has 32-bit precision, meaning the maximum positive value is just over 2 billion. Although R offers various solutions for handling larger numbers, some of these options can lose precision with very large values or are incompatible with other parts of R. Alternatively, Tomasz pointed out that when combining R and Python via reticulate, Python’s integers can be used from within R:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
# Aim: create a large Python NumPy array from R via reticulate
> library(reticulate)
> np <- import("numpy", convert = FALSE)
# Define an integer “i” that is bigger than R’s maximal integer size
# The output “i” is a Python integer object
# The notation 100L is used to signify that the added number should
# be an integer and not a float (which is the default in R)
> i <- r_to_py(.Machine$integer.max) + 100L
> i
2147483747
> class(i)
[1] "python.builtin.int" "python.builtin.object"
# Create a large NumPy array of integers
> a <- np$arange(i)
> a
array([ 0, 1, 2, ..., 2147483744, 2147483745,
2147483746], shape=(2147483747,))
> a$dtype
dtype('int64')
# This integer addition can not be directly performed with R integers
> .Machine$integer.max+100L
[1] NA
Warning message:
In .Machine$integer.max + 100L : NAs produced by integer overflow
> a<-np$arange(.Machine$integer.max+100L)
Warning message:
In .Machine$integer.max + 100L : NAs produced by integer overflow
> a
array([], dtype=int64)
# And since such a large integer type does not exist in R
# a wrong data type will be produced (e.g., float)
> a <- np$arange(2147483747L)
Warning message:
non-integer value 2147483747L qualified with L; using numeric value
> a
array([0.00000000e+00, 1.00000000e+00, 2.00000000e+00, ...,
2.14748374e+09, 2.14748374e+09, 2.14748375e+09],
shape=(2147483747,))
> a$dtype
dtype('float64')
Remainders of R’s 32-bit format
In contrast to Python, R still carries remnants from the 32-bit era, with many common packages still being constrained to 32-bit numbers. This can cause issues when converting from Python to R, as the default object converters may not function properly, requiring specialized tools in R.
For instance, to construct large sparse matrices, a special package spam64 must be used, as other implementations are still 32-bit based. The package spam64 is required whenever the number of entries exceeds 2^31-1, as this is the maximum number of indices (i.e., positive integers) that can be represented in a 32-bit format. This limit can be easily hit, especially with multi-dimensional matrices. For example, modern datasets in the single-cell transcriptomic field contain millions of cells (rows) with over ten thousand genes (columns), surpassing the 32-bit limit.Similarly, while R differentiates between missing values (NA) and impossible values (NaN), such as results of division by zero, NumPy uses a single missing value type (numpy.nan). Starting from R’s NA objects may yield unexpected outcomes in Python, depending on how these values are represented within Python.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
>>> import rpy2.robjects as ro
>>> import numpy as np
# Create an integer vector with NA values in R
>>> ro.r('a<-c(NA,1L)')
>>> ro.r('print(a)')
[1] NA 1
# When such a vector is transported to Python a special NA_integer_ class
# is used to represent R’s NA values
>>> a=ro.r('a')
>>> a
<rpy2.robjects.vectors.IntVector object at 0x119bf3200> [RTYPES.INTSXP]
R classes: ('integer',)
[NA_integer_, 1]
# The class NA_integer_ is interpreted as an integer rather than
# an undefined value within NumPy functions
>>> np.nanmin(a)
np.int32(-2147483648) # Would expect to return 1
# Converting the R type object to NumPy for comparison
>>> a=np.array(a)
>>> a
array([-2147483648, 1], dtype=int32)
Furthermore, custom converters may sometimes be needed. When implementing them, it is important to consider whether the produced object can be converted back into the original language (i.e., make a round-trip). In some cases, information may be lost or altered during conversion. As a result, when the object is reverted to the original language, the results of functions applied to it may be inconsistent or the execution may fail.
For example, to enable round-trips of NaN and NA values between R and Python, reticulate in Python uses different bit patterns to represent nan values that come from R’s NA and NaN. However, unlike R, Python does not handle the two differently by default.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
> library(reticulate)
> np <- import("numpy", convert = FALSE)
# NA and NaN values are represented as nan in Python
> a_r=c(NA,NaN,NA,NaN)
> a_py=np$array(a_r)
> print(a_py)
array([nan, nan, nan, nan])
# The original identity of nan values can be deduced from the bit patterns
# that differ for nan values originating from NA and NaN
> py_to_r(a_py)
[1] NA NaN NA NaN
> a_py$view(np$uint64)
array([9218868437227407266, 9221120237041090560, 9218868437227407266,
9221120237041090560], dtype=uint64)
In some situations, converting the entire object is unnecessary (Figure 5). For example, single-cell transcriptomic data is frequently organized in complex classes containing data, rich metadata, and various results from intermediate analyses. However, to perform differential expression analysis in R, only expression data and metadata related to the design matrix are required. This means that only two data frames or matrices need to be converted, which can be easily managed by the native converters in rpy2 and reticulate. Accordingly, Philipp emphasized that if the conversion fails, developers should inform users which parts of the object could not be converted. If these parts are unnecessary, the user can simply remove them from the object before conversion. This is also preferred to silently omitting inconvertible data parts, as it may lead to unexpected results downstream.
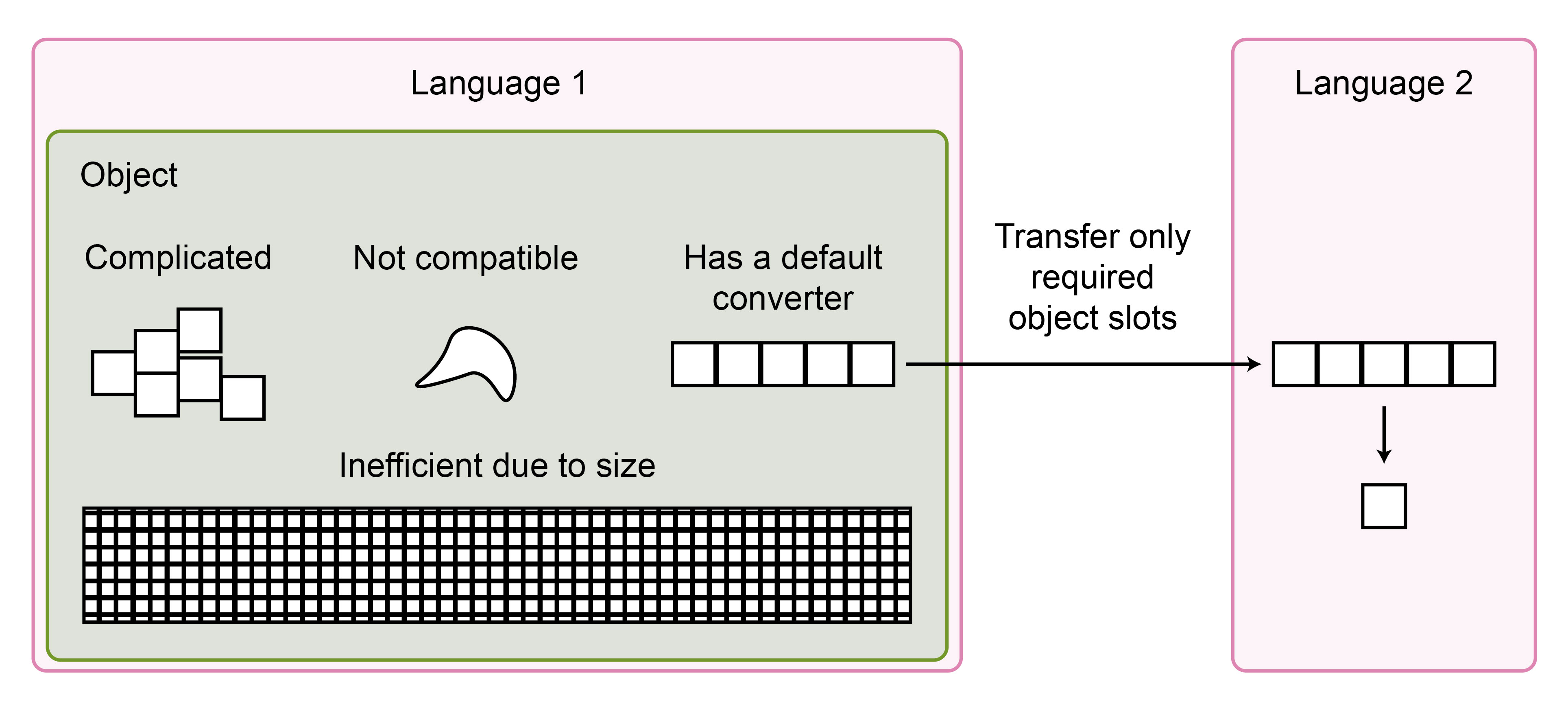 Figure 5: To perform analysis in another language, not all object’s slots may need to be transferred into the other language. Pre-processing of the object can ease conversion.
Figure 5: To perform analysis in another language, not all object’s slots may need to be transferred into the other language. Pre-processing of the object can ease conversion.
Workarounds for accessing data
Lastly, it is important to remember that not only the objects but also the functions may differ between the two languages. Thus, performing similar operations in R and Python might yield different results.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
# Example of corresponding operations with distinct outcomes in R and Python
# Inequality operations on nan values
>>> import rpy2.robjects as ro
>>> import rpy2.robjects.numpy2ri
>>> rpy2.robjects.numpy2ri.activate()
# Inequality operations on undefined values in R return undefined values
>>> ro.r('a<-c(NaN,NA,0)')
>>> ro.r('print(a>0)')
[1] NA NA FALSE
# Inequality operations on undefined values in Python return False
>>> a=ro.r('a') # Use the same vector as above in R
>>> a
array([nan, nan, 0.])
>>> a>0
array([False, False, False])
# Transport inequality evaluation results from Python back to R
>>> ro.globalenv['a_py']=a>0
>>> ro.r('print(a_py)')
[1] FALSE FALSE FALSE
Efficient conversion is no conversion
When evaluating the efficiency of our workflow, it is important to consider which operations are more efficient in each language and how intensive the conversion is. Namely, when converting objects between R and Python, a new copy of the object is created in the target language. This slows down execution and increases memory use.
Therefore, it is best to avoid conversion whenever possible. In reticulate, this can be achieved by using the convert = FALSE flag. For example, we can import a Python package in R with disabled conversion. If this package is then used to create an object in Python and assign it to an R variable, the variable will reference the original Python object rather than an equivalent object in R7 (Figure 6). This is useful when working with large objects in Python that do not require manipulation in R. Instead, only a final summarised result may be subsequently analysed in R.
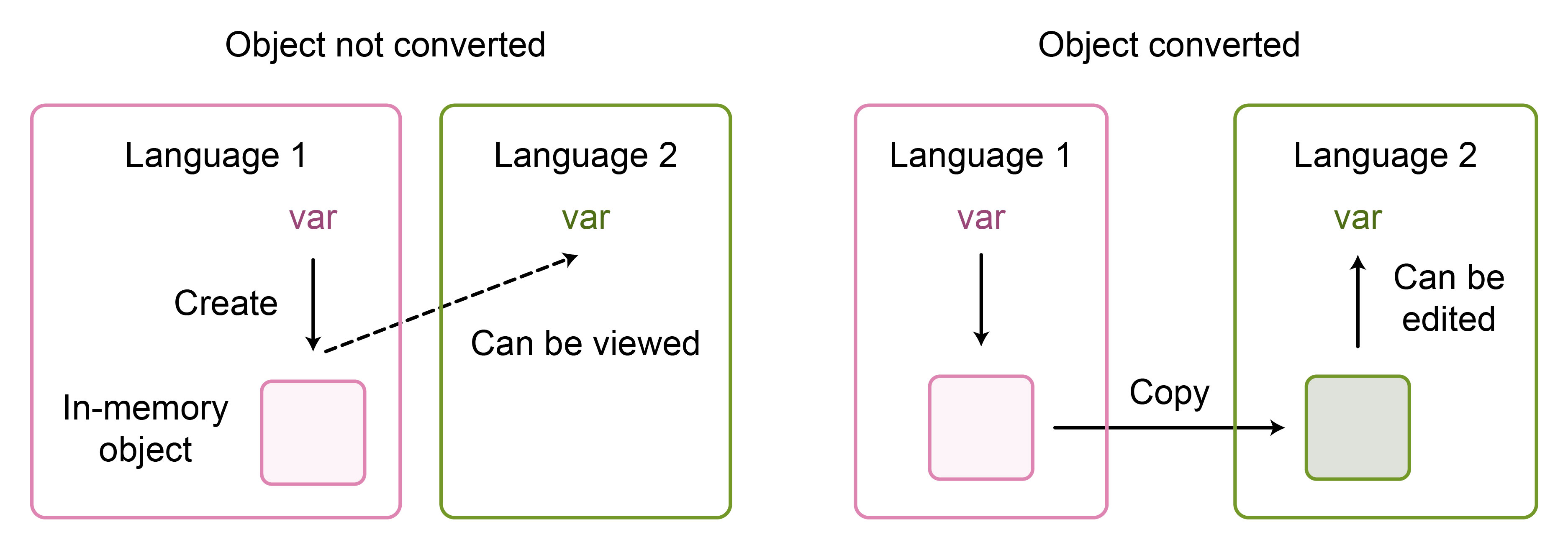 Figure 6: Comparison of converting an object between R and Python (right) and only viewing it in the other language (left). The non-converted objects may still be edited from within the second language via calls to functions from the first language.
Figure 6: Comparison of converting an object between R and Python (right) and only viewing it in the other language (left). The non-converted objects may still be edited from within the second language via calls to functions from the first language.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
# When using reticulate, if Python objects are created with a package that
# uses conversion, they will be converted into R objects.
# In contrast, if no conversion is used, they will remain Python objects.
# Create NumPy array with conversion
> np<-import("numpy",convert=TRUE)
# The object is automatically converted to an R array
> a<-np$array(c(1,2))
> class(a)
[1] "array"
# Create NumPy array without conversion
> np<-import("numpy",convert=FALSE)
# The object remains NumPy array
> a<-np$array(c(1,2))
> class(a)
[1] "numpy.ndarray" "python.builtin.object"
Alternatively, frameworks that share in-memory data between Python and R can be used, such as Arrow. Since less copying is involved, this reduces memory usage and the time required to transfer objects between the two languages8.
Details on sharing memory between R and Python
To achieve truly efficient conversion, low-level computer science concepts must be considered. Although this goes beyond the typical use cases for reticulate and rpy2, it remains an interesting topic to explore.
For instance, copying arrays between R and Python is more efficient if the same memory layout (i.e., column-major ordering) is used in both. This allows contiguous data blocks to be copied directly without rearranging them.
Furthermore, a shared memory layout could theoretically allow R and Python to share a single in-memory object instead of copying it. However, this is complicated in practice as both languages operate under the assumption that they own their memory. For instance, if Python loses its reference to an object (e.g., all variables pointing to it are no longer in scope), it will notify the operating system that it can free up the memory and potentially overwrite it with different data. Subsequently, an error would occur if R tries to access the former object location, which no longer holds the relevant data. Thus, memory is by default not shared between R and Python.
From propagating errors to propagating error messages
When working in two languages at once, it is important to receive meaningful error messages from both sides. Tomasz mentioned that this was one of the key enhancements introduced to reticulate in recent years. Now, a complete error report is displayed irrespective of the language from which the initial error originated. The users can click through the error report to inspect the underlying Python and R code responsible for the error.
Under the hood, the implementation of this feature was rather complex. In short, the developers had to figure out how to effectively catch Python errors within R.
To grasp how the solution works, it is helpful to first understand error propagation. Whenever one function calls another function, a new execution “frame” gets added to the “call stack”. Each frame holds information about the function, such as its local variables, and the location in the code to which execution will return upon function completion. If an exception occurs, it propagates up the stack until caught. This process is known as “stack unwinding”, and as it happens, each traversed frame is removed to release resources.
This process is straightforward when contained within a single programming language. However, when multiple languages are interleaved, the original language might be unable to properly catch and handle exceptions from the other language. Consequently, the error stack might be printed directly to the console and, in the worst case, it may not be fully displayed, complicating debugging. Additionally, as the Python stack has already fully unwound by the time it has reached R code, it ceases to exist and can not be inspected with debugging tools in R (Figure 7).
Reticulate resolved this by converting Python Exceptions to R errors (Figure 7). This provides two main benefits: First, Python exceptions can be caught and handled within R as native R errors. Second, the exception stack can be accessed from within R and used for interactive debugging. Thus, the R IDE can nicely format error messages, including clickable links to the code location of individual frames.
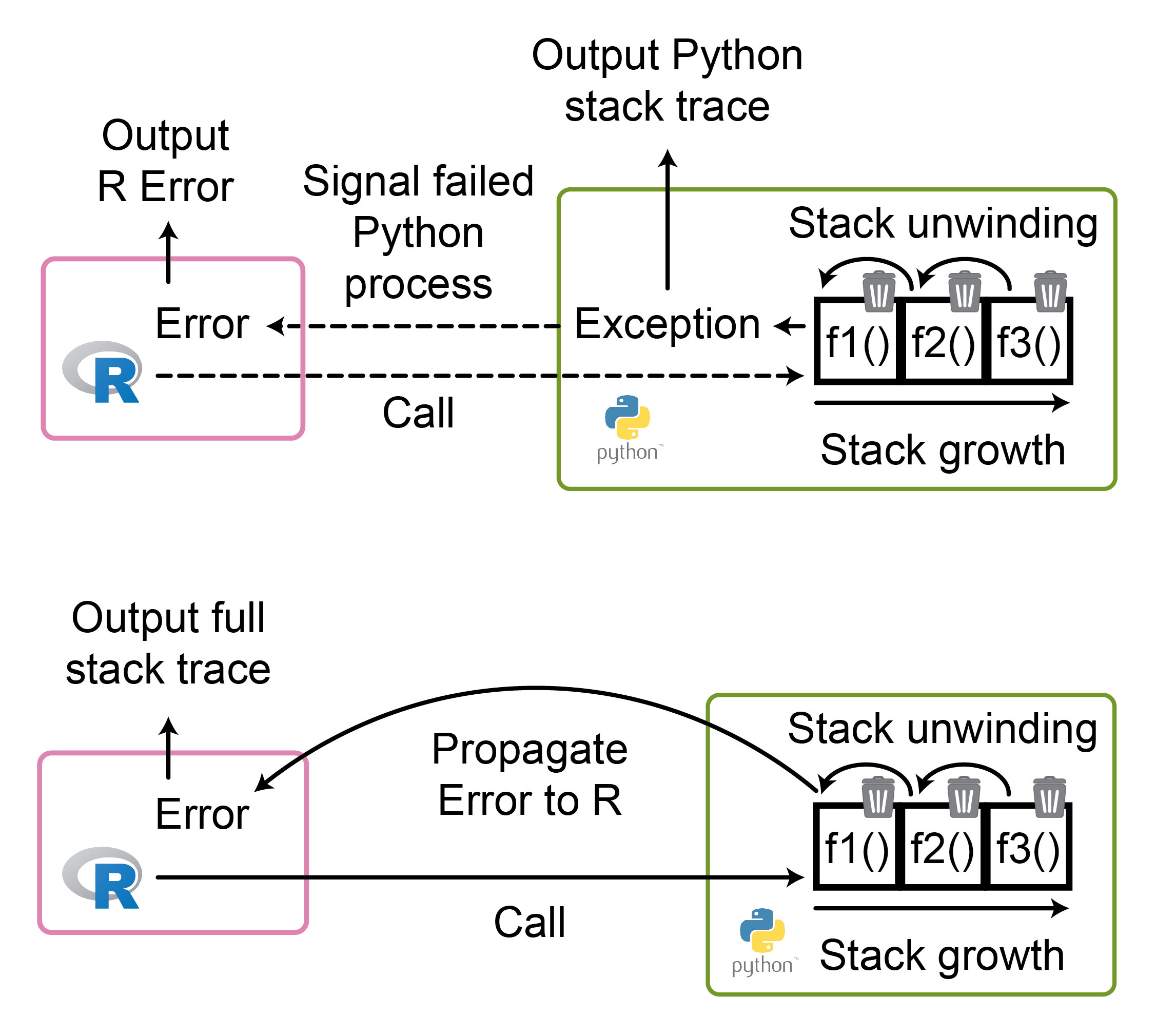 Figure 7: The difference between throwing an Exception in Python (top) and propagating it as an Error to R (bottom) when calling Python from R.
Figure 7: The difference between throwing an Exception in Python (top) and propagating it as an Error to R (bottom) when calling Python from R.
Managing processes
Reticulate is designed to make executing interleaved R and Python commands feel just like executing commands within each language individually. For example, it guarantees that Python execution has completed before new R calls can be made. Moreover, it displays Python terminal outputs in real-time instead of waiting for the Python process to finish9.
However, to achieve this behavior, the main Python thread always remains occupied. Subsequently, applications initiated on other threads may not be able to run, even if no other Python commands are being executed (Figure 8). Namely, with the main thread occupied by reticulate, other threads are blocked by Python’s global interpreter lock (GIL). The GIL ensures thread safety by allowing only one thread to execute Python code at a time, preventing multiple threads from modifying the same object simultaneously.
Luckily, reticulate has established a mechanism allowing Python threads to execute in the background. This facilitates diverse use cases, such as responsive user interfaces, where background threads handle computations without freezing the interface, or data preprocessing in the background, allowing the main thread to concurrently perform model training.
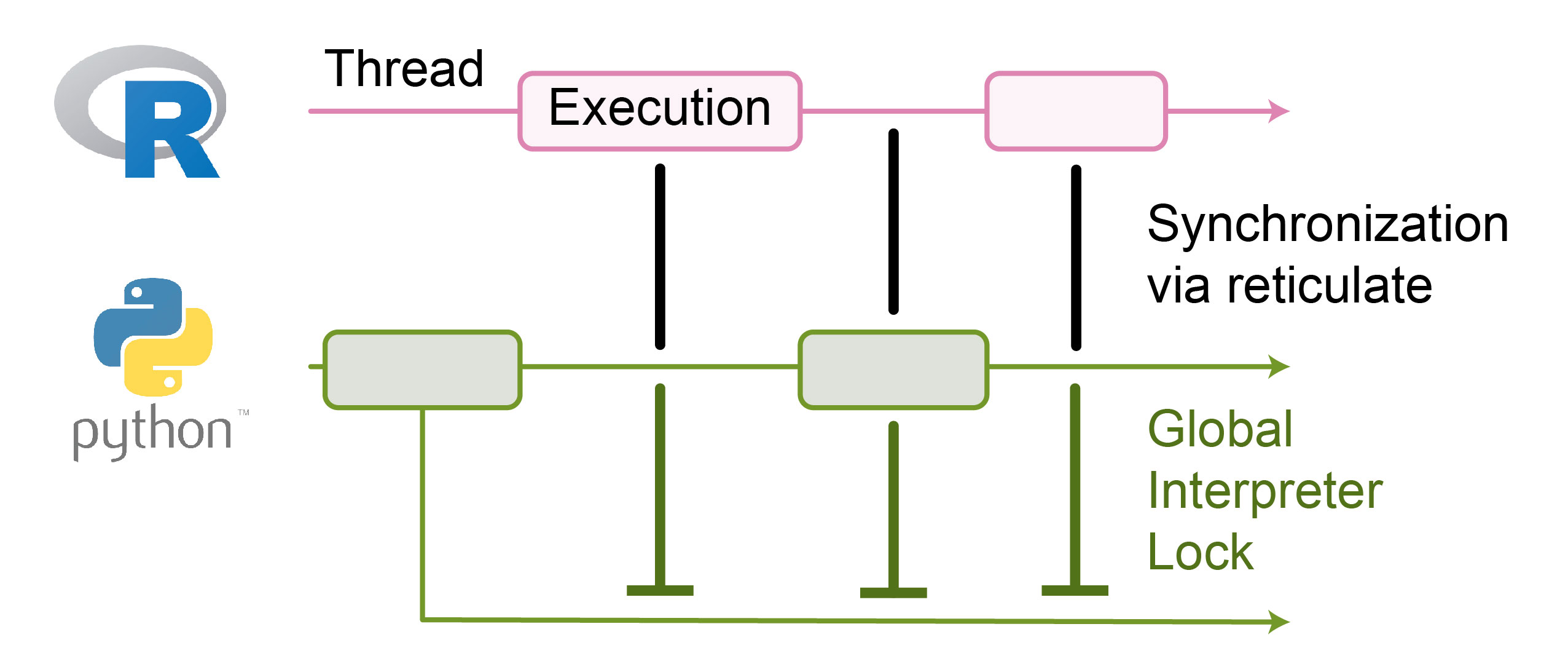 Figure 8: In the past, the global interpreter lock blocked the execution of other Python threads when the main thread was occupied via reticulate. The reticulate’s thread management was resolved to enable parallel execution of multiple Python threads.
Figure 8: In the past, the global interpreter lock blocked the execution of other Python threads when the main thread was occupied via reticulate. The reticulate’s thread management was resolved to enable parallel execution of multiple Python threads.
An example of avoiding multithreading deadlocks
Key takeaways
While interoperability between R and Python is not the main modus operandi for most R and Python users, it is definitely something that more data scientists should be aware of. It easily becomes an indispensable asset if interoperability tools are combined with some considerations on how to best approach it.
Footnotes
In Python, frameworks for out-of-core computation rely on file formats that enable rapid data loading, such as Zarr and HDF5. Besides being able to load individual chunks of data, they also write data to disk in a manner that resembles in-memory storage, thereby minimizing the number of operations required for data retrieval. A popular package for out-of-core computation with these file formats is Dask, which further speeds up computation by parallelizing the processing of individual data chunks. Similar alternatives in R include DelayedArray and Arrow. ↩︎
Interleaving Python and R for data manipulation can be inefficient if objects are continuously copied between the two languages. As described in later sections, this may be addressed with delayed conversion in reticulate. ↩︎ ↩︎2
Apart from seaborn, several other Python alternatives facilitate data visualization, such as Vega-Altair and HoloViews. ↩︎
Bioconductor is a well-known repository for redistributing R packages. It only accepts packages that meet a certain code quality standard and ensures that released packages are compatible with one another and specific R versions. Consequently, Bioconductor packages are more reliable than Python packages distributed via PyPI, which do not need to pass such quality checks. Another popular R repository is CRAN, which also conducts some automatic checks whenever a new package version is submitted. However, Philipp noted that in practice, the quality guarantees of CRAN packages are closer to PyPI than Bioconductor. ↩︎
To accelerate environment setup, uv caches previously downloaded packages and dependency resolution strategies. Additionally, it supports dedicated environments that can be directly reused later. ↩︎
Further examples illustrating differences between R and Python that impact object conversion can be found in reticulate’s documentation. ↩︎
For details on creating Python objects with reticulate without converting them to R, see this tutorial. ↩︎
For a practical introduction to Arrow, see, for example, this blog post. ↩︎
This situation resembles the common Python
subprocessissue, where terminal outputs are only shown after the process concludes. To overcome this, the output buffer must be regularly flushed. ↩︎
