The data curation mindset
Look before you leap!
When it comes to data curation, jumping headfirst into coding is not a good idea. In practice, a better starting point is to sketch out a solid plan.
Most machine learning and data science projects begin with an “exploratory” phase, where different solutions are tried out hands-on. Data curation, however, follows a different workflow: the project planning stage is where exploration takes place. Unlike in computational research, exploration centers on discussions and a manual resource overview, with minimal coding involved. In fact, the bulk of data extraction that follows thereafter resembles the final stages of writing a research paper, where the research plan has already been laid out. In this blog post, we discussed how to approach this mind-shift with Anna Schaar1, Adrian Mirza, and Ada Fang.
Overview of the discussed publications and workshops
Anna was leading the data collection and evaluation of the single-cell foundation model Nicheformer (Tejada-Lapuerta and Schaar, 2025). One of the biggest challenges for the curation team was to finish extracting information from diverse data formats within the planned timeframe.
Adrian was leading the ChemPile project (Mirza, 2025), which collected datasets for training LLMs specialised in chemistry. Due to the project's scale, it was important for the entire team to agree on how the data should be collected and processed.
Ada was one of the organizers of the NeurIPS 2025 dataset proposal competition. Their goal was to identify emerging scientific domains that would benefit from dedicated machine learning models, along with the datasets required for their training. Each contestant submitted a brief proposal outlining the creation of a dataset designed to enable the use of machine learning to address a specific scientific problem.
This blog post highlights topics that got less focus in the two publications and on the workshop website. If you are more generally interested in dataset curation, I recommend reading the original papers, as many topics are skipped here. Furthermore, this post does not aim to provide a comprehensive overview of the field and instead focuses on highlights from our conversations.
While this blog post is based on discussions with Anna1, Ada, and Adrian, I often omit their names for the sake of readability.
Sections list
- Organizing data curation
- Weighting manual curation quality against LLM speed
- Working in a curation team
- NeurIPS workshops as a platform for new projects
- Curation from start to end
- What to collect and package?
- How to define evaluation tasks?
Organizing data curation
Data curation is not typically seen as the “most fun” of activities. Finding where manual effort adds the greatest value, and how to make it more enjoyable, is therefore important for every curation project.
Weighting manual curation quality against LLM speed
When it comes to collecting single-cell datasets, Anna is not yet convinced that a primarily LLM-based workflow is feasible, given the limitations of current agentic capabilities. The process is complex even for human curators and often involves unexpected obstacles that are not straightforward to resolve. - Different data shards from each dataset are commonly scattered across multiple locations (Figure 1) and deposited in non-standard formats (see “The horrors of curating single-cell datasets”). This makes it difficult to define a clear task for the agent in advance. - And even when the task is clear, the data often contains outliers that are not handled well, as illustrated by Adrian’s experience with optical character recognition (OCR, see “Papers are easy to read for humans, but not for LLMs”). Therefore, it is important to decide where human curation is essential and which steps can be automated.
The horrors of curating single-cell datasets
In simplified terms, single-cell datasets typically consist of expression information for a set of genes (i.e., features) measured in each cell (i.e., sample), along with associated metadata. The metadata may be defined on the cell level (e.g., cell type annotation), or on the dataset level (e.g., how the expression was preprocessed). Although some existing formats try to standardize the collection of this information, at least in part (e.g., the cellxgene schema), datasets are rarely deposited accordingly.
For example, most datasets are still deposited on GEO, which does not specify a standard format for single-cell data. Consequently, expression data appears in many forms, ranging from one matrix per cell to one per subject (e.g., person or animal), or one per entire dataset. Moreover, there is no standard matrix file format; common examples include sparse matrices and CSVs.
When it comes to metadata, the situation is even worse. Often, important pieces are missing or deposited in a completely different location (Figure 1), meaning researchers must track them down from the paper and its supplements. - Sometimes, the data is not even in a format that can be read programmatically. For example, tables are often provided as images, requiring manual curation or OCR to extract the information.
The metadata nomenclature also tends to be dataset-specific. What is more, often it is not even enough to look up synonyms in the ontology. For example, data may contain abbreviations that are explained only in the manuscript. Furthermore, animal datasets are occasionally annotated with human gene names rather than those of the source species, which means additional investigation is needed to determine why certain names can not be recognized.
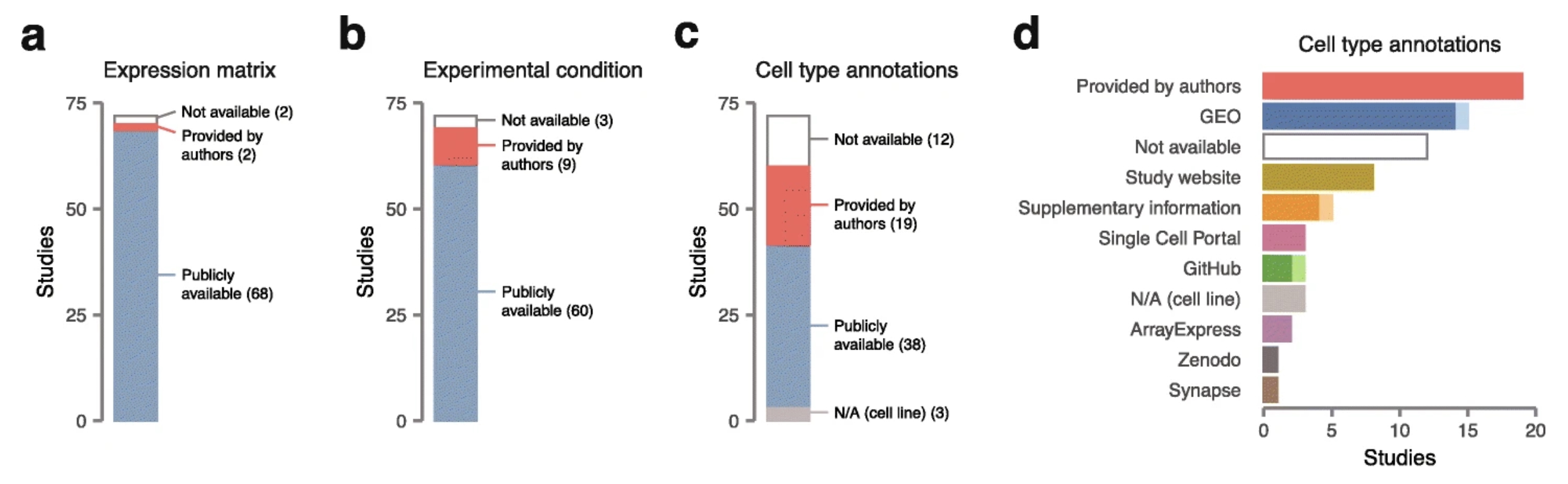 Figure 1: Unstandardized data deposition practices in the single-cell field. The image shows data availability for 72 published scRNA-seq datasets, as provided by Skinnider (2021). Availability of (a) processed gene expression matrices, (b) sample-level metadata (e.g., treatment vs. control), and (c) cell-level metadata (i.e., cell type annotation). (d) Sources from which cell type annotations were obtained, with light shades representing datasets for which multiple manual processing steps were required to obtain cell types. The image was adapted from Figure 1 of (Skinnider, 2021), which is published under CC BY 4.0 license.
Figure 1: Unstandardized data deposition practices in the single-cell field. The image shows data availability for 72 published scRNA-seq datasets, as provided by Skinnider (2021). Availability of (a) processed gene expression matrices, (b) sample-level metadata (e.g., treatment vs. control), and (c) cell-level metadata (i.e., cell type annotation). (d) Sources from which cell type annotations were obtained, with light shades representing datasets for which multiple manual processing steps were required to obtain cell types. The image was adapted from Figure 1 of (Skinnider, 2021), which is published under CC BY 4.0 license.
Papers are easy to read for humans, but not for LLMs
When it comes to human curation, Adrian also pointed out the importance of deciding how tasks should be distributed among team members. In his experience, it is best to organize curation by topic, allowing domain expertise to be applied where it is most needed, rather than by data type or resource (e.g., papers versus code). - While organizing by data type might make workflows more efficient, since team members can specialize, leveraging domain expertise is more valuable.
Working in a curation team
Curating datasets for large-scale models usually requires a team. Navigating teamwork is thus as important as defining the technical focus.
Both Anna and Adrian emphasized that it is key to have both good organization as well as a motivating atmosphere. They listed a few factors that can help support this:
- Providing tools that ease repetitive work. This goes beyond helper scripts2 and also includes ways to make the work more enjoyable. Both Anna and Adrian suggested using an app that allows team members to complete small tasks whenever they have a few spare minutes. For example, the ChemPile team built a web app for validating curation quality (Figure 2), which gradually presented a few examples to each team member.
- Allowing team members to find their own ways of working. The motivation behind this is not only to accommodate different work styles but also to offer the opportunity for trying out new approaches, thus making the work less mundane. Still, it is important that this freedom does not undermine the standards crucial for model training.
- Following a clear work plan - and having someone actively oversee it. Otherwise, joint progress can quickly stall, as Anna noted. Adrian also observed that collaborating with a team from a single research institute made progress easier, as both the schedule and the curation standards were simpler to coordinate. In addition, for personal time management, Anna recommended reserving a fixed curation time block. This ensures that curation tasks are not waved aside - but also do not overload you.
- Having well-functioning communication, with dedicated people to consult for each topic. However, Adrian pointed out that having the option to ask should not become a crutch for lacking documentation. - This will backfire once people move on and are no longer available.
- Working reproducibly to ease future adjustments. For example, ChemPile data extraction code and accompanying documentation were deposited on GitHub through a peer-reviewed process. In addition, the code should be modular enough to allow easy corrections within isolated data subsets without affecting the rest of the data.
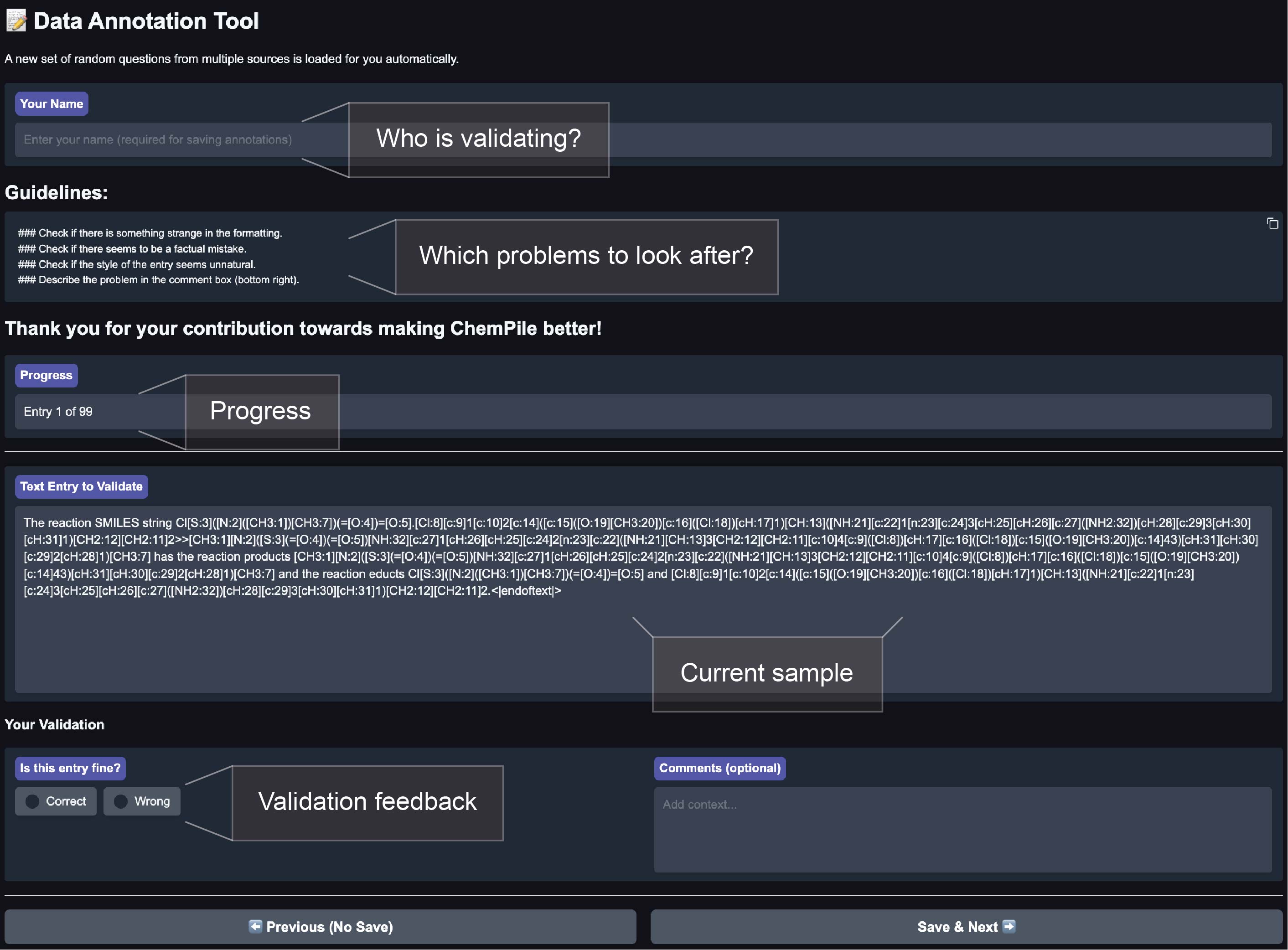 Figure 2: App for validation of curated data used by the ChemPile team. By flipping through examples from each dataset, curation errors were identified.
Figure 2: App for validation of curated data used by the ChemPile team. By flipping through examples from each dataset, curation errors were identified.
The skillset and resources to build foundation models - academia or industry?
Building true foundation models requires substantial financial resources and long-term commitment. Anna pointed out that such large-scale investments are not the primary purpose of academia, speculating that future foundation models for biology are likely to emerge from industry.
Academic labs also often lack the expertise to train models efficiently. Even the most talented PhD students rarely have prior experience training foundation models, likely leading to increased consumption of compute resources.
Last but not least, training a foundation model requires considerable discipline and a well-aligned team. While this is not impossible in academia, it conflicts with the primary purpose of doctoral programs, which is personal development. - If the priority is model quality and robustness, even long, mundane tasks must be completed meticulously from start to finish. Consequently, dedicating time during doctoral studies to such work may not be optimal, as the focus should be on exploring new things.
Besides this, Anna noted that fine-tuning foundation models for specific datasets or tasks that are not well captured by the model may often not be the optimal choice. Since fine-tuning can be expensive, developing smaller, specialized models can often be a worthwhile alternative.
NeurIPS workshops as a platform for new projects
NeurIPS workshops generally mirror the format of the main conference, with papers and talks. However, Ada believes that they could be used as a unique venue for experimenting with new ideas - not just in terms of publication topics but also in how the workshops are carried out.
Their dataset proposal competition3 already moves in this direction. By adopting an unconventional manuscript format, it raised awareness of application domains that are often overlooked in the machine learning community - a common bottleneck in developing machine learning solutions, as Ada noted. As a result, the value of the competition extended beyond new ideas for efficient dataset collection4, also revealing which scientific domains would benefit from machine learning models in the future5.
Another enticing alternative would be to use the workshop event to work jointly on a shared problem. NeurIPS workshops are a great venue for this, as they can be attended by a large group of experts who are already on site for the main conference.
Potential workshop topics
Organizing a new format type will, inevitably, come with some hiccups. For example, Ada pointed out that it was hard to evaluate dataset proposals. They differ substantially from the typical NeurIPS manuscript, which usually focuses on a specific methodological contribution supported by empirical results. In contrast, reviewing proposals requires assessing the feasibility and innovation of the proposed approach, which calls for a different set of evaluation criteria. Additionally, because most reviewers have a machine learning background, it was more difficult to assess the feasibility and potential value of ideas rooted in new application domains.
Balancing participants’ time investment against the quality of workshop outcomes
Curation from start to end
Curation should start and finish with questions about how the data will ultimately be used. For example, Adrian explained that their data curation mirrored the LLM training workflow, encompassing datasets for all stages from pre-training to fine-tuning6. At the same time, curators should anticipate and facilitate unforeseen ways in which the data might be reused.
What to collect and package?
Defining what constitutes “useful” data is challenging. In practice, there is no definitive answer, as it depends on the task at hand. For this reason, Anna emphasized the importance of depositing all of the available information when publishing a dataset, even if some of it was not used in the original project. With a similar mindset, the ChemPile team designed their dataset as a modular platform, allowing others to decide which parts to use and in what way.
A crucial fact often forgotten is that not every large dataset is ML-ready. In the NeurIPS challenge, many of the submissions focused on high-throughput platforms. However, having millions of samples that cover only a narrow portion of the possible data diversity is not enough. Anna pointed out that when scraping biological data, this is sadly common. Certain tissue types are heavily overrepresented in public databases (Figure 3) because they are easier to obtain (e.g., blood vs. eye) or of higher interest (e.g., lung cancer vs. rare diseases). Consequently, adding yet another million healthy blood cells may contribute little to improving the general performance of single-cell foundation models.
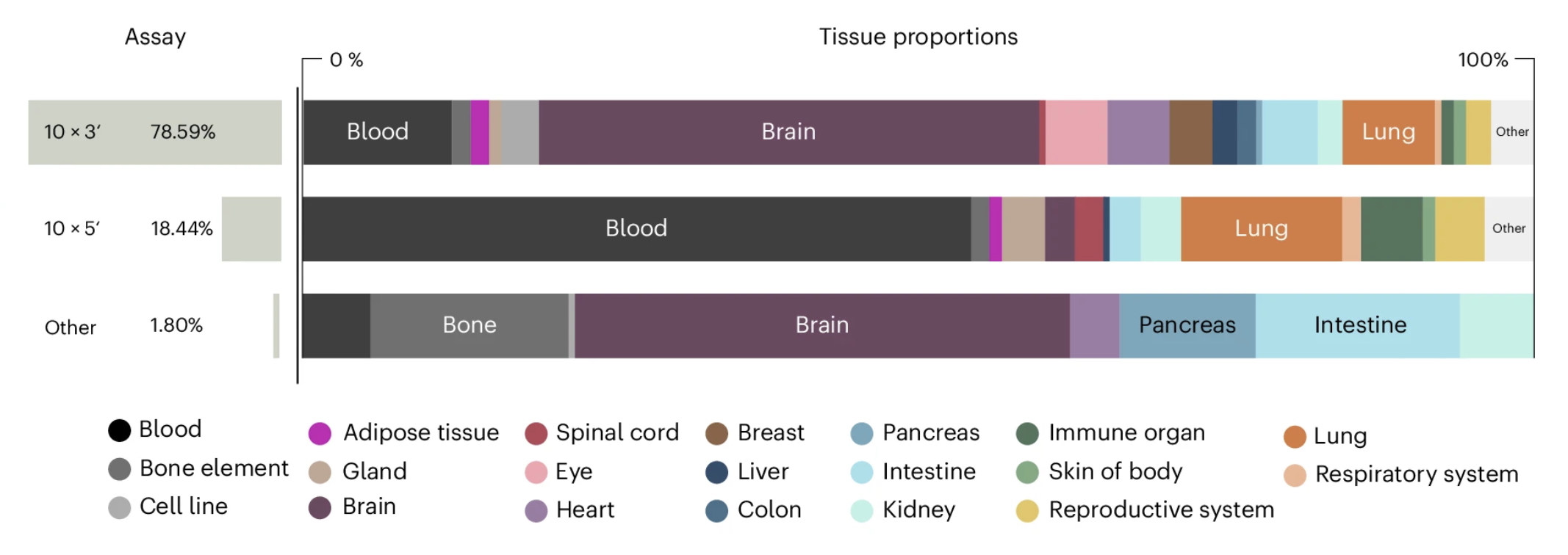 Figure 3: Unbalanced tissue type proportions in published single-cell datasets. Tissue proportions are shown across three types of measurement assays. The figure was adapted from Figure 2a of (Tejada-Lapuerta and Schaar, 2025), which is published under CC BY 4.0 license.
Figure 3: Unbalanced tissue type proportions in published single-cell datasets. Tissue proportions are shown across three types of measurement assays. The figure was adapted from Figure 2a of (Tejada-Lapuerta and Schaar, 2025), which is published under CC BY 4.0 license.
Another important yet often overlooked component is the license. When asked about community requests regarding ChemPile, this was the main point Adrian highlighted. Because they did not consider licenses at the start, the team later had to revisit each dataset to determine its license status in order to make ChemPile ready for use (Figure 4).
 Figure 4: Overview of different license restrictions. In the Creative Commons licensing framework, different licensing restrictions can be combined to provide the desired level of content protection. Note that Creative Commons lists NC as more permissive than ND, and I have adapted the order based on my personal experience.
Figure 4: Overview of different license restrictions. In the Creative Commons licensing framework, different licensing restrictions can be combined to provide the desired level of content protection. Note that Creative Commons lists NC as more permissive than ND, and I have adapted the order based on my personal experience.
When collecting data for a specific task, such as model training, it is essential to agree on the data requirements from the outset. Without it, Anna mentioned, data curation may never be completed, and model training cannot begin. - While it is always possible to add more metadata later, doing so often means starting almost from scratch. A good example are the messy single-cell depositions (described under “The horrors of curating single-cell datasets”), where adding more technical metadata typically requires re-reading the paper.
We need to increase the focus on technical metadata
Is ontology data valuable for LLM training?
In ChemPile, the team deliberately left out ontology data, as curating it was considered an unnecessary effort. LLMs are highly capable of learning relationships between terms directly from the text, whereas ontologies often lag behind the latest research terminology. Ontologies also miss many synonym variations, such as abbreviations, which are nevertheless commonly used in publications. Thus, to understand the text, LLMs need to learn these relationships directly from the data anyway.
Besides defining what information would be valuable, it is also important to consider how long the curation will take. Both Anna and Adrian mentioned that they were constantly running against the clock. Thus, for Nicheformer, they calculated how much time could be spent per dataset to reach the desired dataset size by their chosen data-freeze date. This helped set realistic expectations on how detailed the metadata extraction could be. Similarly, Adrian mentioned that during data validation, they often chose to simply drop isolated erroneous entries rather than spend additional time resolving them to include every last piece. Large-scale data curation, therefore, requires a different mindset compared to smaller projects, where every data point may be cherished.
Finally, Anna stressed that a carefully curated, high-quality dataset is one of the most valuable resources for any computational researcher. Thus, when effort is invested in curration, the value of sharing it should not be overlooked. Adrian added that the data should be structured so that it is both easily understandable and reusable in a custom - and potentially unforeseen - way.
Challenges of depositing single-cell data across omic layers and scales
- For patient data, which is duplicated across all cells of a patient, storing metadata at the cell rather than the subject level is inefficient.
- There is no clear convention for storing tissue images and representing relationships between image patches and cells (e.g., cell type annotations or expression).
How to define evaluation tasks?
When constructing large-scale data resources, it is important to define which subsets should be set aside for evaluation. With Anna and Adrian, we discussed a couple of points that are important to consider:
- Diversity within each evaluation task. For example, to evaluate the annotation of cell niches in a specific tissue, the test data should come from multiple patients or animals, not just one or two. - Even if each subject contributes thousands of sample cells, they will all carry the same subject and dataset-specific biases.
- Different levels of generalisation, with both in-distribution and out-of-distribution splits. For some modalities, like chemical structures, approaches for creating such splits are well established. However, in single-cell space, the concept of data similarity remains unclear. For example, if the same dataset is subjected to different preprocessing, it may appear substantially different, despite having identical biology. This can lead to data leakage when using multiple composite datasets (e.g., atlases), which incorporate multiple, potentially overlapping, primary datasets that were processed differently within each atlas.
- Different types of tasks become especially important for foundation models, which aim to serve as a general tool for anyone in the field. For example, the evaluations in single-cell community focus on cell type (i.e., class) annotation and batch (i.e., domain) correction tasks, stemming from earlier works on representation learning for domain correction. However, this does not cover emerging applications and data modalities. For example, in spatial data, it is not only relevant to know the general type of a cell (e.g., blood or brain cell), but also the finer-grained biological variation that stems from its niche within the tissue.
Ideally, benchmarks should be defined on the community level. As an example, Anna highlighted the CZI jamboree on single-cell foundation model benchmarking. Without such community-driven standards, it is easy for model developers to cherry-pick datasets that favor their model rather than focusing on tasks of broad interest. That said, even when everyone agrees on the benchmarking tasks, these tasks are not immune to being over-optimized for, quickly losing their ability to reflect general model performance.
Key takeaways
Data collection requires careful coordination from the very beginning, ensuring that questions of both usability and resource requirements are thoughtfully addressed. And since there are many mundane tasks to get through, it is just as important to keep the team spirit alive.
References
- Blecher, et al. Nougat: Neural Optical Understanding for Academic Documents. ICLR (2024).
- Mirza, et al. ChemPile: A 250GB Diverse and Curated Dataset for Chemical Foundation Models. arXiv (2025).
- Skinnider, et al. Enabling reproducible re-analysis of single-cell data. Genome Biology (2021).
- Tejada-Lapuerta and Schaar, et al. Nicheformer: a foundation model for single-cell and spatial omics. Nature Methods (2025).
Footnotes
This post represents Anna’s personal opinions and does not reflect those of her current employer. ↩︎ ↩︎2
Helper scripts can be, for example, used to validate that the curated data matches the defined schema. As an example, see the cellxgene validator. ↩︎
The accepted dataset proposals are published at OpenReview within track 2. ↩︎
The dataset generation strategies of the winning proposals were diverse and included: de novo measurement, computational simulations that require expert knowledge and substantial resources, human labeling, community contribution of internal datasets, and curation of existing scattered public sources. ↩︎
The winning proposals tackled challenges across different scientific domains: marine biodiversity tracking based on acoustic signals, automated parsing of engineering machine diagrams (e.g., car parts), cell segmentation in common histological (i.e., tissue) images, ozone modeling within the troposphere (i.e., lowest atmospheric layer), and modeling of kinetic mechanisms within catalyst-aided chemical reactions. ↩︎
They collected general materials, such as code, papers, and lectures, for pre-training, as well as various types of datasets for specific fine-tuning tasks, including tabular data for property prediction and Stack Exchange data for enhancing reasoning capabilities. ↩︎
