Data Sci. updates
The most unexpected and/or useful data science bits I have learned in the past weeks.
This page serves as an archive of these updates.
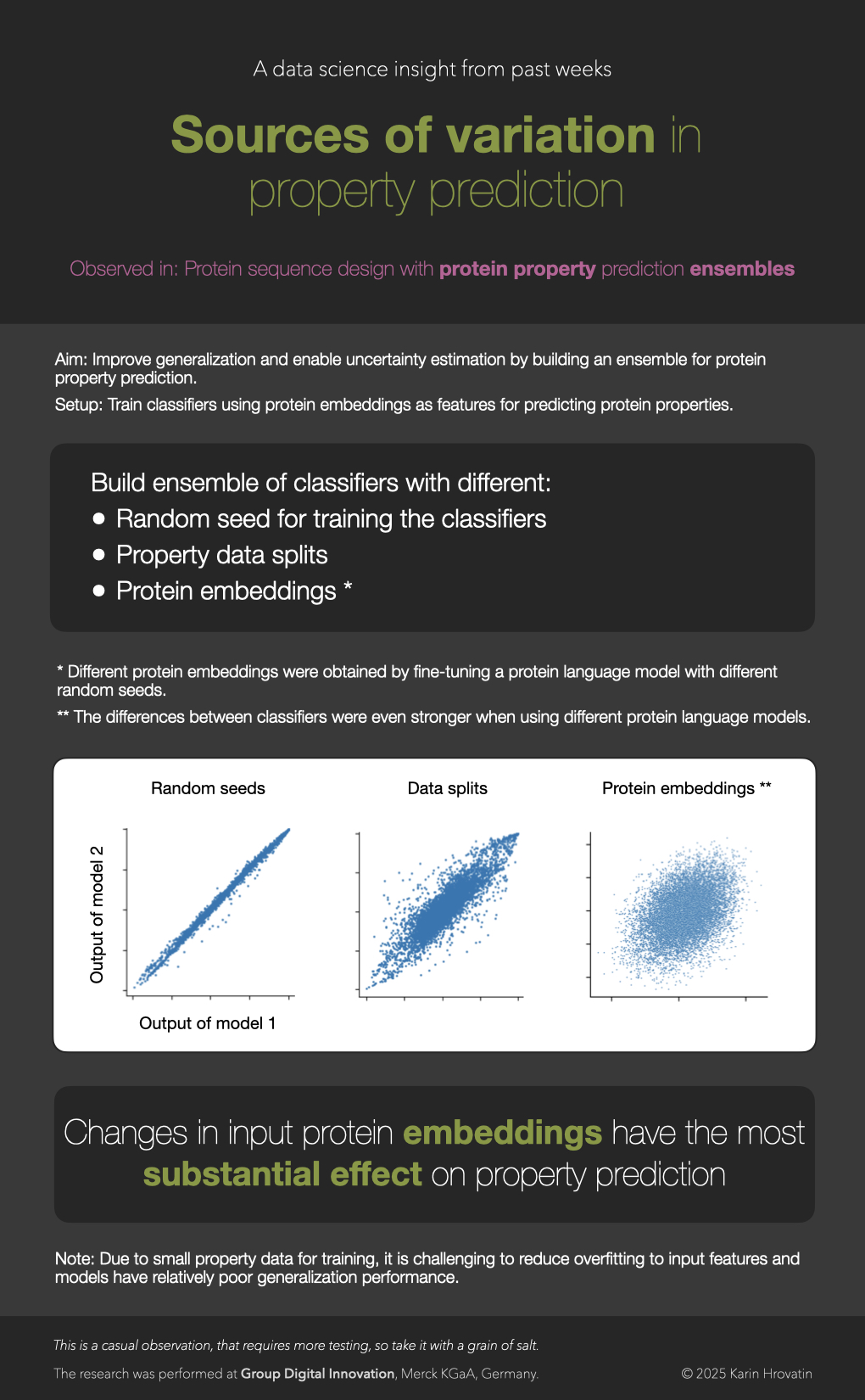
Sources of variation
The strongest source of variation in protein property prediction (in a low-data regime) seem to be protein representations, rather than classical ensemble-building techniques (e.g., different random seeds or data splits for training).
Biases in uncertainty estimates
The relatively uncertainty of regions away from training data can be underestimated. One of the reasons behind this may be a combination of underfitting in regions away from data, which may result in more uniform predictions across models within an ensemble, and overfitting in regions close to the data, leading to higher variability across ensemble members.
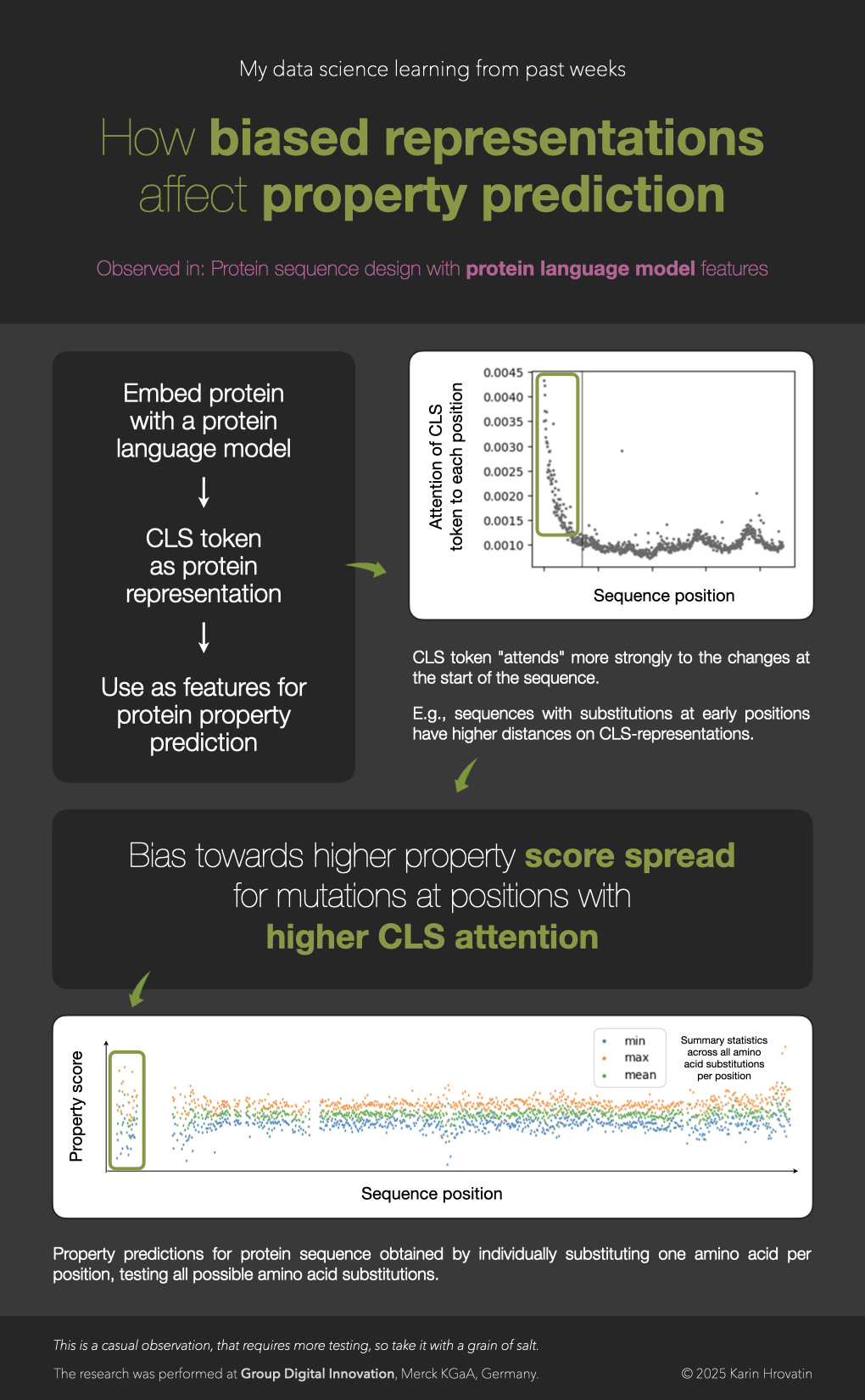
CLS token representation biases
When using CLS token as protein representations, the mutations in certain regions of protein sequence may be over-prioritized.